Introduction
The goal of codemapper is to simplify working with clinical codes for research using electronic health records. The workflow is as follows:
- Create a local resource containing lookup and mapping tables for various clinical codings systems (e.g. ICD10 and Read codes)
- Build clinical code lists for conditions of interest by querying this resource
This vignette demonstrates the above using dummy data included with the package. You can try out the steps either locally by installing codemapper on your own machine, or online by clicking on the following link to RStudio Cloud1 and navigating to this Rmd file in the ‘vignettes’ directory:
Also included are functions for mapping between different clinical coding systems, and using CALIBER code lists(Kuan et al. 2019) and Phecodes(Denny, Bastarache, and Roden 2016; Wu et al. 2019) with UK Biobank data. See vignettes vignette('map_codes') vignette('caliber') and vignette('phecodes') for further information.
Build a local clinical codes lookup and mappings resource
The first step is to create a local database containing lookup and mapping tables for various clinical coding systems using build_all_lkps_maps().
By default this will download the following resources:
UK Biobank resource 592 (Clinical coding classification systems and maps)
UK Biobank data codings file
Phecode lookup and mapping files (for ICD9 and ICD10 to phecode)
The tables are imported into R, reformatted, and stored as a named list of data frames:
# build dummy all_lkps_maps resource (supressing warning messages)
all_lkps_maps_dummy <- build_all_lkps_maps_dummy()
# view first few rows of ICD10 lookup table
head(all_lkps_maps_dummy$icd10_lkp)
#> # A tibble: 6 × 13
#> .rowid ICD10_CODE ALT_CODE USAGE USAGE_UK DESCRIPTION MODIFIER_4 MODIFIER_5
#> <int> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 1 A00 A00 DEFAULT 3 Cholera NA NA
#> 2 2 A00.0 A000 DEFAULT 3 Cholera due… NA NA
#> 3 3 A00.1 A001 DEFAULT 3 Cholera due… NA NA
#> 4 4 A00.9 A009 DEFAULT 3 Cholera, un… NA NA
#> 5 5 A01.0 A010 DEFAULT 3 Typhoid fev… NA NA
#> 6 6 A02 A02 DEFAULT 3 Other salmo… NA NA
#> # … with 5 more variables: QUALIFIERS <chr>, GENDER_MASK <chr>, MIN_AGE <chr>,
#> # MAX_AGE <chr>, TREE_DESCRIPTION <chr>Many of the functions in this package will require this object (supplied to argument all_lkps_maps).
Instead of re-running build_all_lkps_maps() in each new R session, the output can be saved to a SQLite database file:
# write to SQLite database file
db_path <- suppressMessages(all_lkps_maps_to_db(all_lkps_maps = all_lkps_maps_dummy,
db_path = tempfile()))Connect to this and create a named list of dbplyr::tbl_dbi2 objects:
# connect to SQLite database
con <- DBI::dbConnect(RSQLite::SQLite(), db_path)
# create named list of tbl_db objects
all_lkps_maps_dummy_db <- ukbwranglr::db_tables_to_list(con)
# view first few rows of ICD10 lookup table
head(all_lkps_maps_dummy_db$icd10_lkp)
#> # Source: SQL [6 x 13]
#> # Database: sqlite 3.38.5 [/tmp/Rtmp8l1oBv/filee812b61abf1]
#> .rowid ICD10_CODE ALT_CODE USAGE USAGE_UK DESCRIPTION MODIFIER_4 MODIFIER_5
#> <int> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 1 A00 A00 DEFAULT 3 Cholera NA NA
#> 2 2 A00.0 A000 DEFAULT 3 Cholera due… NA NA
#> 3 3 A00.1 A001 DEFAULT 3 Cholera due… NA NA
#> 4 4 A00.9 A009 DEFAULT 3 Cholera, un… NA NA
#> 5 5 A01.0 A010 DEFAULT 3 Typhoid fev… NA NA
#> 6 6 A02 A02 DEFAULT 3 Other salmo… NA NA
#> # … with 5 more variables: QUALIFIERS <chr>, GENDER_MASK <chr>, MIN_AGE <chr>,
#> # MAX_AGE <chr>, TREE_DESCRIPTION <chr>Either will work with the functions described in the remainder of this vignette, but the SQLite database option is recommended. For convenience, record the path to your SQLite database in a .Renviron file:
ALL_LKPS_MAPS_DB=/PATH/TO/all_lkps_maps.dbAssuming you are using a RStudio project, this will set an environmental variable called ALL_LKPS_MAPS_DB when you start a new R session. Functions with an all_lkps_maps argument will automatically search for this variable and attempt to connect with the database file at that path, meaning you will not need to repeatedly type this in your self.
Build a clinical code list
Explore codes
Codes may be explored with:
```r
lookup_codes(codes = c("E10", "E11"),
code_type = "icd10",
all_lkps_maps = all_lkps_maps_dummy_db)
#>
[38;5;246m# A tibble: 2 × 3
[39m
#> code description code_type
#>
[3m
[38;5;246m<chr>
[39m
[23m
[3m
[38;5;246m<chr>
[39m
[23m
[3m
[38;5;246m<chr>
[39m
[23m
#>
[38;5;250m1
[39m E10 Type 1 diabetes mellitus icd10
#>
[38;5;250m2
[39m E11 Type 2 diabetes mellitus icd10
``````r
codes_starting_with(codes = "E1",
code_type = "icd10",
all_lkps_maps = all_lkps_maps_dummy_db)
#>
[38;5;246m# A tibble: 55 × 3
[39m
#> code description code_type
#>
[3m
[38;5;246m<chr>
[39m
[23m
[3m
[38;5;246m<chr>
[39m
[23m
[3m
[38;5;246m<chr>
[39m
[23m
#>
[38;5;250m 1
[39m E10 Type 1 diabetes mellitus icd10
#>
[38;5;250m 2
[39m E100 Type 1 diabetes mellitus With coma icd10
#>
[38;5;250m 3
[39m E101 Type 1 diabetes mellitus With ketoacidosis icd10
#>
[38;5;250m 4
[39m E102 Type 1 diabetes mellitus With renal complications icd10
#>
[38;5;250m 5
[39m E103 Type 1 diabetes mellitus With ophthalmic complications icd10
#>
[38;5;250m 6
[39m E104 Type 1 diabetes mellitus With neurological complications icd10
#>
[38;5;250m 7
[39m E105 Type 1 diabetes mellitus With peripheral circulatory complic… icd10
#>
[38;5;250m 8
[39m E106 Type 1 diabetes mellitus With other specified complications icd10
#>
[38;5;250m 9
[39m E107 Type 1 diabetes mellitus With multiple complications icd10
#>
[38;5;250m10
[39m E108 Type 1 diabetes mellitus With unspecified complications icd10
#>
[38;5;246m# … with 45 more rows
[39m
```code_descriptions_like():
```r
code_descriptions_like(
reg_expr = "cyst",
code_type = "icd10",
all_lkps_maps = all_lkps_maps_dummy_db
)
#>
[38;5;246m# A tibble: 2 × 3
[39m
#> code description code_type
#>
[3m
[38;5;246m<chr>
[39m
[23m
[3m
[38;5;246m<chr>
[39m
[23m
[3m
[38;5;246m<chr>
[39m
[23m
#>
[38;5;250m1
[39m L721 Trichilemmal cyst icd10
#>
[38;5;250m2
[39m N330 Tuberculous cystitis icd10
```R Shiny app
Use RunCodelistBuilder() to launch a R Shiny app for building a clinical code list.3 The aim is to develop a broad search strategy that captures all clinincal codes that might possibly be included, then download the search results and manually select and sub-categorise the final set of codes:
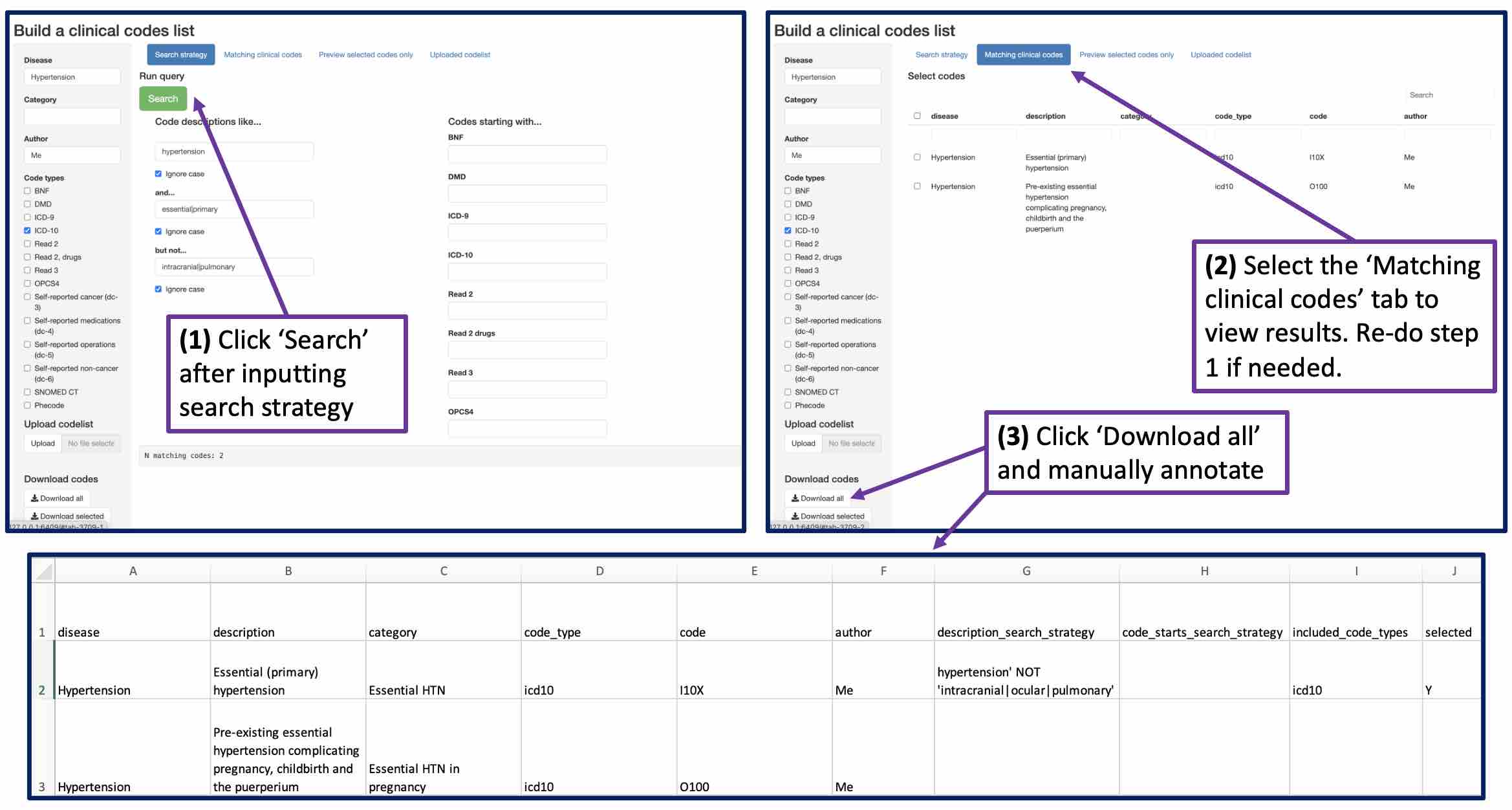
Build a clinical code list in R Shiny
Microsoft excel auto-formatting can cause problems with certain codes e.g. Read 3 ‘.7944’ (Creation of permanent gastrostomy) may be reformatted to ‘7944’. Ideally use a different text editor.
Denny, Joshua C., Lisa Bastarache, and Dan M. Roden. 2016. “Phenome-Wide Association Studies as a Tool to Advance Precision Medicine.” Annual Review of Genomics and Human Genetics 17 (August): 353–73. https://doi.org/10.1146/annurev-genom-090314-024956.
Kuan, Valerie, Spiros Denaxas, Arturo Gonzalez-Izquierdo, Kenan Direk, Osman Bhatti, Shanaz Husain, Shailen Sutaria, et al. 2019. “A Chronological Map of 308 Physical and Mental Health Conditions from 4 Million Individuals in the English National Health Service.” The Lancet. Digital Health 1 (2): e63–e77. https://doi.org/10.1016/S2589-7500(19)30012-3.
Wu, Patrick, Aliya Gifford, Xiangrui Meng, Xue Li, Harry Campbell, Tim Varley, Juan Zhao, et al. 2019. “Mapping ICD-10 and ICD-10-CM Codes to Phecodes: Workflow Development and Initial Evaluation.” JMIR Medical Informatics 7 (4): e14325. https://doi.org/10.2196/14325.
You will be asked to sign up for a free account if you do not have one already.↩︎
If you have not used SQL with R before, I recommend reading the Get started vignette from the dbplyr package.↩︎
This is still quite experimental, but should hopefully work for the basic workflow described here.↩︎
